1. INTRODUCTION
Biochemistry, sometimes called biological chemistry, is the study of chemical processes in living organisms, including, but not limited to, living matter. Biochemistry governs all living organisms and living processes. By controlling information flow through biochemical signaling and the flow of chemical energy through metabolism, biochemical processes give rise to the complexity of life.
Around two dozen of the 94 naturally-occurring chemical elements are essential to various kinds of biological life. Just six elements—carbon, hydrogen, nitrogen, oxygen, calcium, and phosphorus—make up almost 99% of the mass of a human body.
Essential Elements (By weight) in Protoplasm
1. Oxygen – 63% 4. Nitrogen – 3.0%
2. Carbon – 20% 5. Calcium – 2.5%
3. Hydrogen – 10% 6. Phosphorus – 1.4%
A Comparison of Elements Present in Non-living and Living Matter*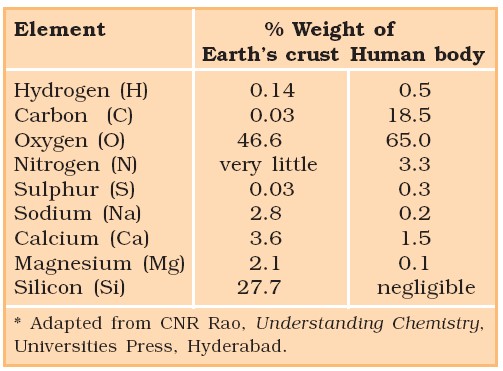
2. THE IMPORTANT COMPOUNDS IN CELLULAR POOL
Biomolecules
- All the carbon compounds that we get from living tissue, can be called biomolecules. It includes both organic and inorganic substances.
- Organic compounds can be analyzed by the following method
Average Composition of Cells
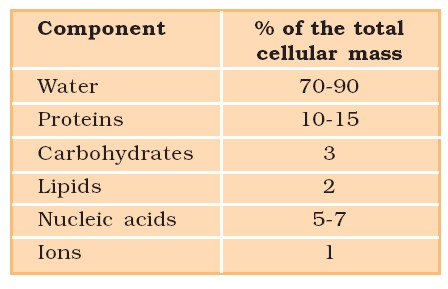
- Grind a living tissue (a piece of vegetable or liver) in trichloroacetic acid in a mortar with a pestle to obtain a thick slurry
- Filter this slurry of living tissue through a cheese cloth or cotton to obtain two fractions: Filtrate (acid-soluble pool) and retentate (acid-insoluble fraction). The acid soluble fraction represents the micro biomolecules of cytoplasm while acid insoluble fraction represents macro biomolecules and organelles.
Inorganic elements and compounds can be analyzed by the following procedure.
- Weigh a small amount of a living tissue and note down its wet weight.
- Dry the living tissue by evaporating its water in an oven and note down its dry weight.
- Burn the dried tissue to oxidize all the organic compounds to obtain ash which contains inorganic compounds of calcium, magnesium, etc. Inorganic compounds like phosphates and sulphates are also found in acid-soluble fraction.
- Ash or acid-soluble fraction is subjected to elemental analysis to know elemental composition of living tissues in the form of carbon, oxygen, hydrogen, chloride sodium (Na+), potassium (K+), calcium (Ca++), magnesium (Mg++), inorganic compounds like NaCl, CaCO3, phosphates (PO33–), sulphtes (SO42–), etc.
Biomicromolecules / Vs Biomacromolecules
| Biomolecules | |
| Acid Soluble Fraction (micromolecules) | Acid insoluble Fraction (macromolecule) |
|
|
* because of smaller size of lipids (few), it can not be considered as strict biomacromolecule.
Metabolites
Small or big compounds (chemicals) which can be isolated from living organisms. They are classified into two basic types.
| Primary Metabolites | Secondary Metabolites |
| These are biomolecules required for basic metabolic processes. i.e., have identifiable role and play known roles in physiological functions. | These are derivatives of primary metabolites. |
| These are found throughout the plant and living organism. | Some Secondary metabolites are found in Plant, fungal and microbial cells. |
| These are part of the basic molecular structure of the cell. | These are not part of the basic molecular structure of the cell. |
| They are highly useful to plant. | They have limited role in plant, or its function is not known. |
Example of secondary metabolites.
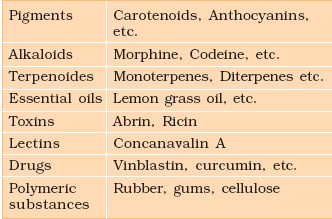
3. CARBOHYDRATES
Carbohydrates are derived polyhydroxy of Aldehyde or ketone.
Classification of Carbohydrates
Monosaccharide
General formula of monosaccharide is Cn(H2O)n where n is the number of C-atom in a monosugar which varies from 3 to 7.
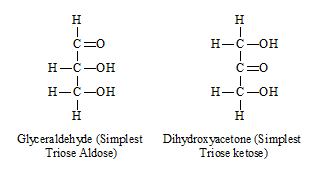
Various monosaccharide are
| No. of C | Aldose | Ketose |
| 3 (C3H6O3) | Glyceraldehyde | Dihydroxyacetone |
| 4 (C4H8O4) | Erythrose | Threose |
| 5 (C5H10O5) | Ribose, deoxyribose, xylose | Ribulose |
| 6 (C6H12O6) | Glucose, Galactose | Fructose |
Structure of Glucose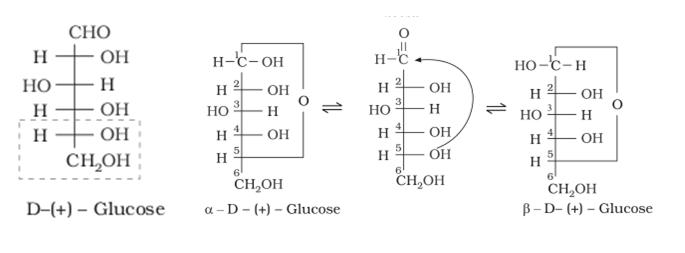
Ring Structure of Glucose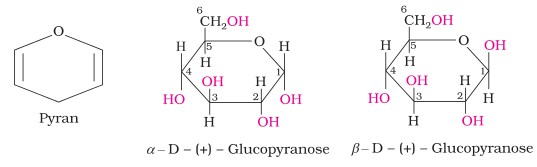
Structure of Fructose
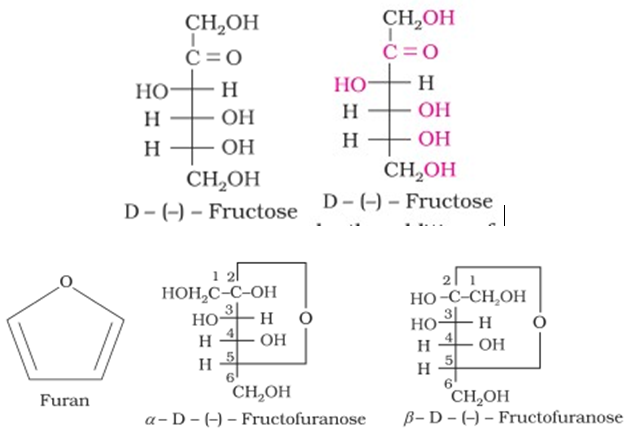
The cyclic structures of two anomers of fructose are represented by Haworth structures as given.
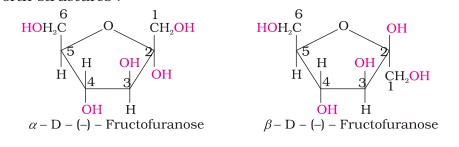
Disaccharides – They contain 2-monosaccharides
(i) Maltose – It is sugar and formed during germination of starchy seeds. It is reducing sugar. It contains 2 α – glucose units (with α-1,4-linkage/glycosidic bond)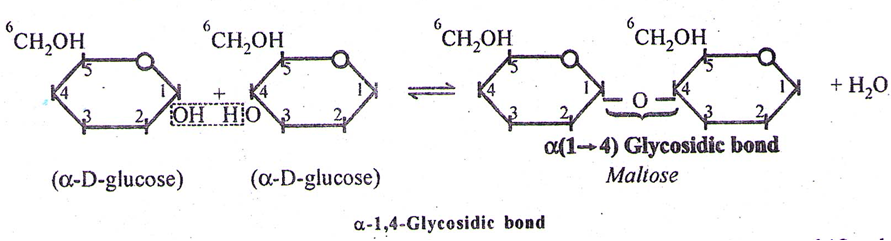
Other disaccharides and its linkages
| Disaccharide | Unit | Unit 2 | Bond |
| Sucrose (table sugar, cane sugar, beet sugar, or saccharose | glucose | fructose | α (1 → 2) |
| Lactose (Milk sugar) | galactose | glucose | β (1 → 4) |
| Maltose | glucose | glucose | α(1 → 4) |
| Trehalose | glucose | glucose | α (1 → 1)α |
| Cellobiose | glucose | glucose | β (1 → 4) |
Trisaccharides – They contains 3 monosaccharides, e.g., Raffinose – 1 glucose + 1 fructose + 1 galactose.
Polysaccharides
- They are formed by joining of Monosaccharides by Glycosidic bonds between 1-4 carbon atoms.
- Depending upon the types of monosaccharides or monomers, polysaccharides may be homopolysaccharides or heteropolysaccharides
| Polysaccharides | |||||
| Structural | Storage | Conjugated or Complex | |||
|
Cellulose
|
Chitin
|
Plant Starch | Animal |
|
|
| Amylose | Amylopectin | Glycogen | |||
|
|
|
|||
- Inulin → storage polysaccharide in plant, a polymer of β-fructose
- In a polysaccharide chain, the right end is called the reducing end and the left end is called the non-reducing end. (Right end must have free Aldehyde or ketone group)
- Starch forms helical secondary structures. In fact, starch can hold I2 molecules in the helical portion. The starch-I2 is blue in colour.
- Cellulose does not contain complex helices and hence cannot hold I2.
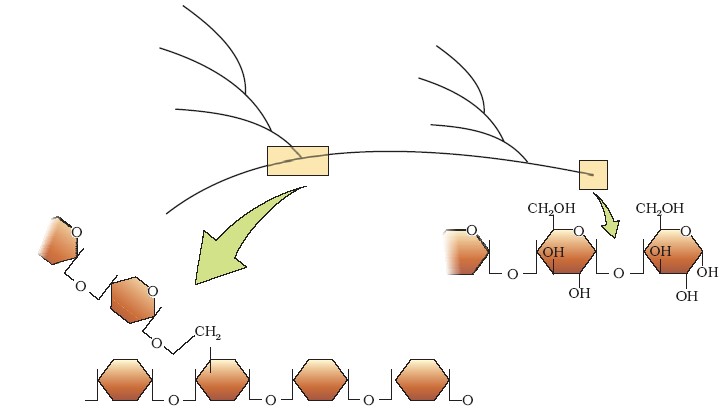
Diagrammatic representation of a portion of glycogen
3. Mucopolysaccharides – e.g., Heparin. Hyaluronic acid, Synovial fluid, Vitreous humor, cell wall in bacteria and Mucilages (Galactose and mannose). They are also present in Bhindi (Lady’s finger) and Isabgol
Carbohydrate Test – Benedict’s test, Fehling’s test.
- All monosaccharide and disaccharide (except sucrose) are reducing sugar.
- Other oligosaccharide and Polysaccharides are non-reducing sugar, due to the absence of free aldehyde or ketone group
4. LIPIDS
- Lipids are non-polymers, micromolecules as well as macromolecule.
- Lipids are ester of fatty acids and glycerol
| Classification of Fatty acid | |
| Saturated FA | Unsaturated FA |
| Ex – Palmitic acid (C16H32O2)
Stearic acid (C18H36O2) Arachidic acid (C20H40O2) |
1. Oleic acid (C18H34O2) (Single double bond) (MUFA)
2. Linoleic acid (C18H32O2) (2 – double bond) 3. Linolenic (C18H30O2) (3 double bond) 4. Arachidonic acid (C20H32O2) (4 double bond) (2, 3, 4 are polyunsaturated fatty acid) (PUFA) |
| Essential FA | Nonessential FA |
| It can not be synthesized in the animal body and must be supplied with food to avoid its deficiency. Ex – Linoleic acid, Linolenic acid, Arachidonic acid |
Can be synthesized in the body tissues so may or may not be present in that diet Ex – Palmitic acid stearic acid, oleic acid |
Human body – fat contains 50% Oleic acid, 25% Palmitic acid, 10% Linoleic acid and 5% Stearic acid.
3 CLASSES – 1. Simple lipids 2. Compound lipids 3. Derived lipids.
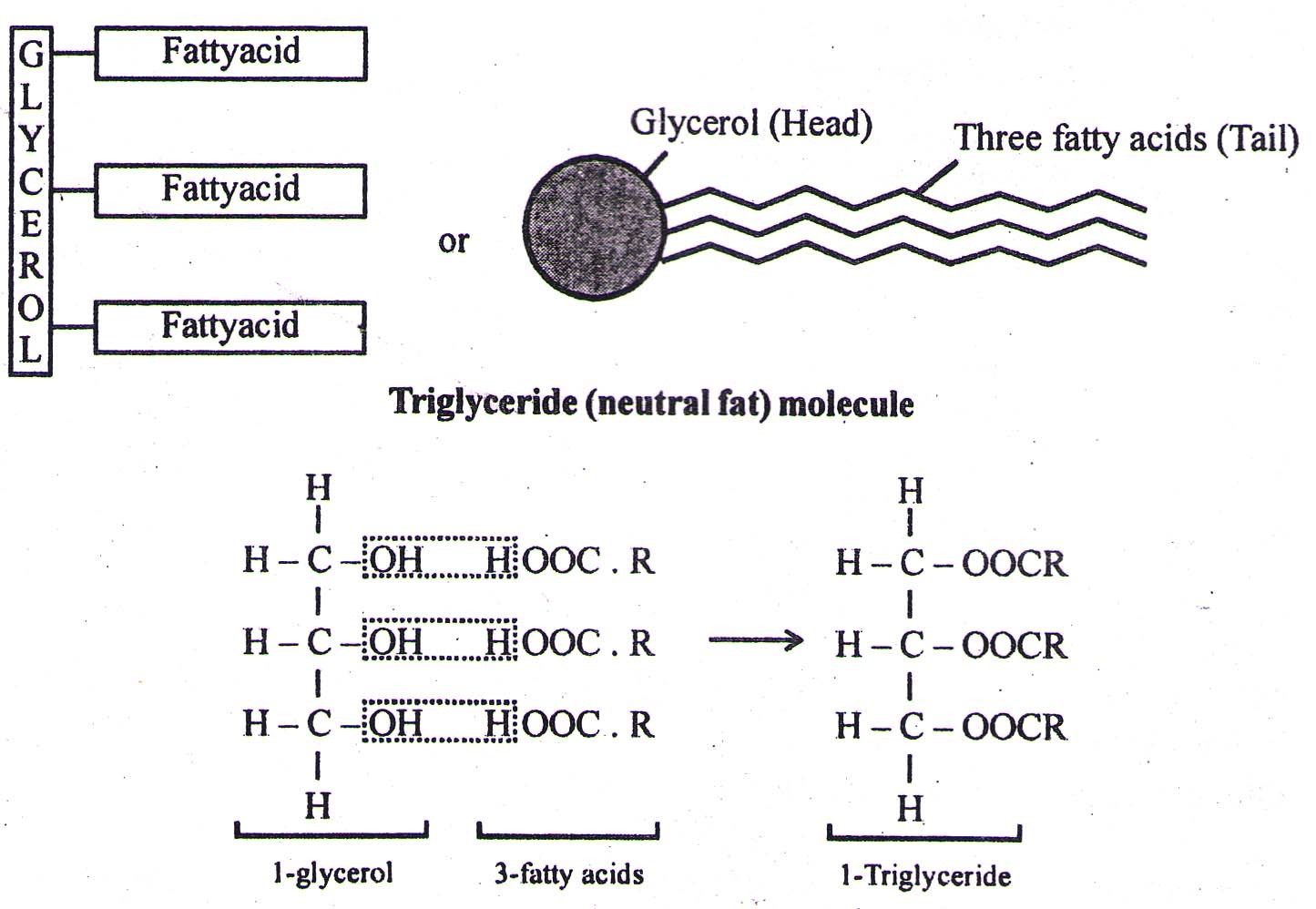
(1) SIMPLE LIPIDS (Homolipids) – They are esters of fatty acids with various alcohols
- 3-fatty acids are attached to a C-atom of glycerol (forming triglyceride) by Ester linkage (a weak bond, which is broken on heating).
- The fats that are liquid at room temp. (20ºC) are called OILS, i.e., Ground nut oil, Mustard oil, Til oil and Safflower oil. The oils contain Unsaturated fatty acids which on hydrogenation form saturated fatty acids or GHEE.
- Oils have lower melting point (e.g., gingelly oil) and hence remain as oil in winters.
- They are all straight chain compounds and are also called as Neutral Fats – e.g. Oil, Butter, Margarine, Ghee etc.
Waxes – e.g. Paraffin, Bee wax, Sebum
- Esters of fatty acids with alcohols, higher than glycerol. (Alcohol in waxes have only one hydroxyl group compared to 3-in glycerol)
(2) COMPOUND LIPIDS
A. Phospholipids – e.g. Lecithin (present in cell membranes)
They are constructed like a neutral fat, except that is place of 3rd fatty acid, there is a phosphate group. The phosphate group with glycerol becomes polar head (Hydrophilic) of the molecule while hydrocarbon chains of fatty acids become the non-polar tails.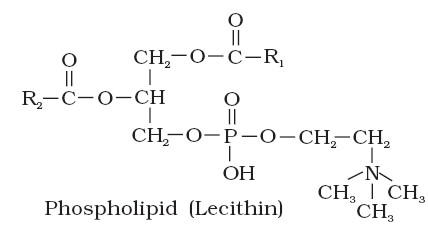
B. sphingolipids – They are also present in cell membranes and are comparable to phospholipids except that sphingosine is present instead of glycerol. Each sphingolipid molecule has a hydrophilic (water soluble) ‘head’ , and one-longer and one-shorter hydrogen tails.(Hydrophobic).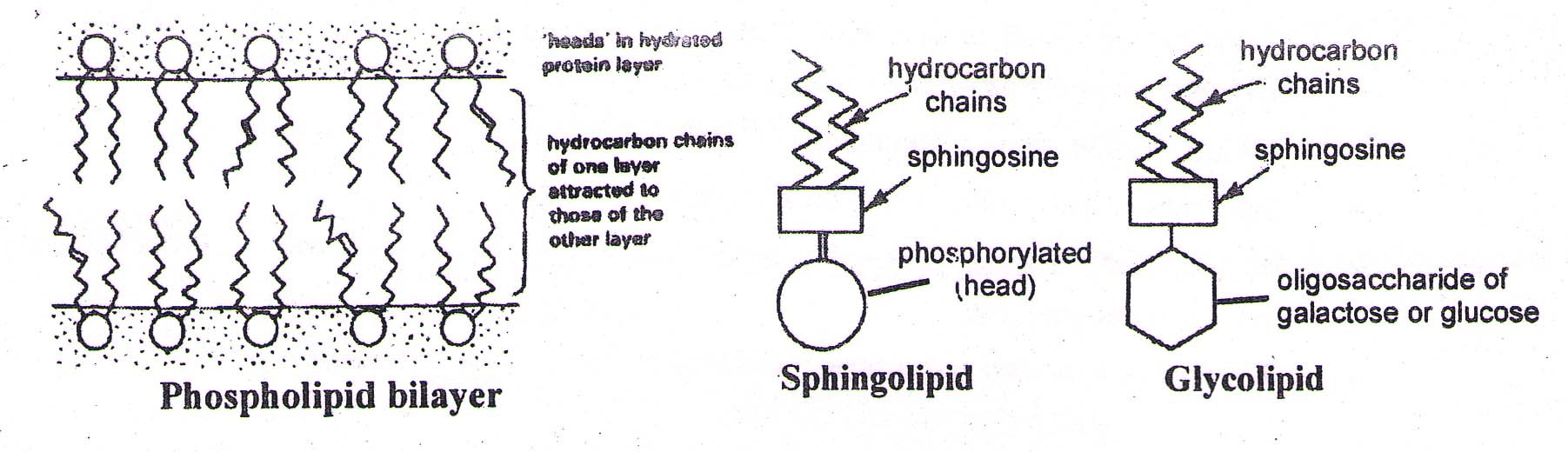
C. Glycolipids – They are like sphingolipids except that oligosaccharide (of glucose / galactose) is bonded to sphingosine instead of the phosphoric acid. The oligosaccharide (head) is water soluble. Hence, their properties are like sphingolipids. eg, A & B antigens in ABO system of blood group.
(3) DERIVED LIPIDS – E.g., Monoglycerides, Diglycerides, Steroids, Sterol Terpenes and Carotenoids etc. They are formed by hydrolysis of fats.
Test for Lipids- (1) Grease Spot Test (2) Sudan III (black) or Sudan red test
Functions-
- Poor conductor of heat (For Insulation)
- Shock absorber
- Food storage (Its primary function)
5. PROTEINS
- Proteins are most diverse chemicals (Macromolecules) in the living organisms.
- Proteins contain C, H, O, N. Some contain S (Sulphur) and P(Phosphorus) also. The structural unit of protein is Amino acid.
- They are polymers and contain more than 100 amino acids, with one of more polypeptide chains.
- They are synthesized in the body from 20-different types of amino acids only.
Amino Acid
- They are digestive end products of proteins
- All contains atleast 1-Amino group and 1-carboxylic group.
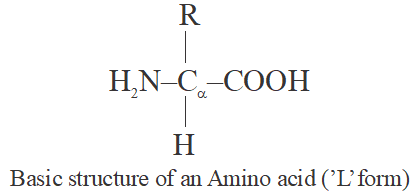
- Most of the amino acids are amphoteric in nature (with one amino group and one carboxylic group).
- Amino acids are organic compounds containing an amino group and an acidic group as substituents on the same carbon i.e., the α-carbon. Hence, they are called α-amino acids. They are substituted methanes.
- The chemical and physical properties of amino acids are essentially of the amino, carboxyl and the R functional groups.
- A particular property of amino acids is the ionizable nature of –NH2 and –COOH groups. Hence in solutions of different pHs, the structure of amino acids change.
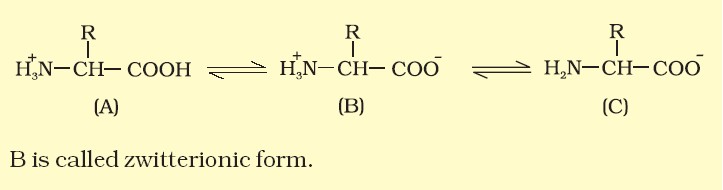
Classification of Amino Acid
| Essential | Nonessential |
| 1. Isoleucine (IIe) 2. Leucine (Leu) 3. Lysine (Lys) 4. Methionine (Met 5. Phenylalanine (Phe) 6. Threonine (Thr) 7. Tryptophan (Trp) 8. Valine (Va) |
1. Alanine (Al) 2. Asparagine (Asn) 3. Aspartic Acid (Asp) 4. Cysteine (Cys) 5. Glutamic Acid (Glu) 6. Glutamine (Gln) 7. Glycine (Gly) 8. Proline (Pro) 9. Serine (Ser) 10. Tyrosine (Tyr) |
| Acidic | Basic | Neutral | |
| 1. Aspartic Acid 2. Glutamic Acid |
1. Arginine 2. Histidine 3. Lysine |
Rest other amino acids are neutral | |
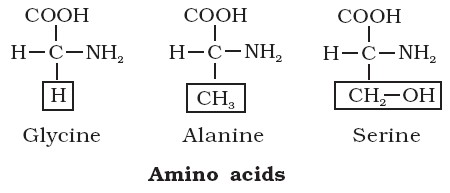
- Glycine is the simplest amino acid, while tryptophan is the most complex one.
- Phenylalanine, Tyrosine and Tryptophan contain benzene ring and so are Aromatic amino acids. (Tyrosine is formed in the body from Phenylalanine).
- Proline, a cyclic amino acid, is also called as Imino acid.
- Serine and Threonine are hydroxyl amino acids.
- The amino acids Cysteine and Methionine contain Sulphur.
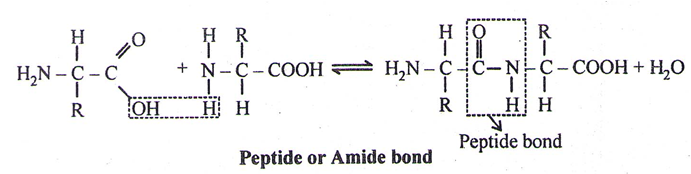
Peptide
- A peptide is imagined as a line, the left end represented by the first amino acid and the right end represented by the last amino acid. The first amino acid is also called as N-terminal amino acid. The last amino acid is called the C-terminal amino acid.
- The different amino acids are attached through – CO-NH bond, called Peptide bond or Amide bond.
Important peptides –
Glutathione – It is a tripeptide (with 3 amino acids) having Glycine, Glutamic acid and Cysteine
Oxytocin and Vasopressin – They have 9-amino acids each
Bradykinin – It has 9-amino acids. It is most potent pain-producing substance which mediates prostaglandins.
Angiotensin – Angiotensin-l with 10 amino acids and Angiotensin – II with 8 amino acids are vasoconstrictors.
Types of Proteins – Simple protein, Conjugated proteins, and Derived proteins
(i) Simple Proteins-
- Composed of amino acids only
- Soluble or insoluble in water
- Simple proteins include Albumins, Globulins, Protamines, Lectins and Scleroproteins. The fibrous proteins (scleroproteins) include collagen and Keratin etc.
| Conjugated Proteins | |
| Glycoproteins | Proteins combined with carbohydrates. E.g., Hormones, FSH, LH, TSH and HCG, Blood group, Serum protein etc. |
| Lipoproteins | Proteins combined with lipids e.g. Cell membrane and chylomicrons, LDL (Low density Lipoproteins) and HDL (High density lipoproteins) |
| Nucleoproteins | Proteins attached to nucleic acids, e.g. Histones in chromosomes (Histones are basic proteins, i.e. rich in basic amino acids – lysine and arginine). |
| Phosphoproteins | Protein containing phosphorous as prosthetic group, e.g. Casein of milk and egg vitelline. |
| Metalloproteins | Proteins with metal ions, e.g. Tyrosinase (copper), Carbonic anhydrase (zinc), Cytochrome, (iron), Insulin (zinc). |
| Chromoproteins | Proteins with pigment of coloured prosthetic group, e.g. flavoproteins (yellow), hemoglobin (red), visual pigment (purple) etc. |
Derived Proteins
They are produced during hydrolysis of proteins, eg. Proteoses and Peptones.
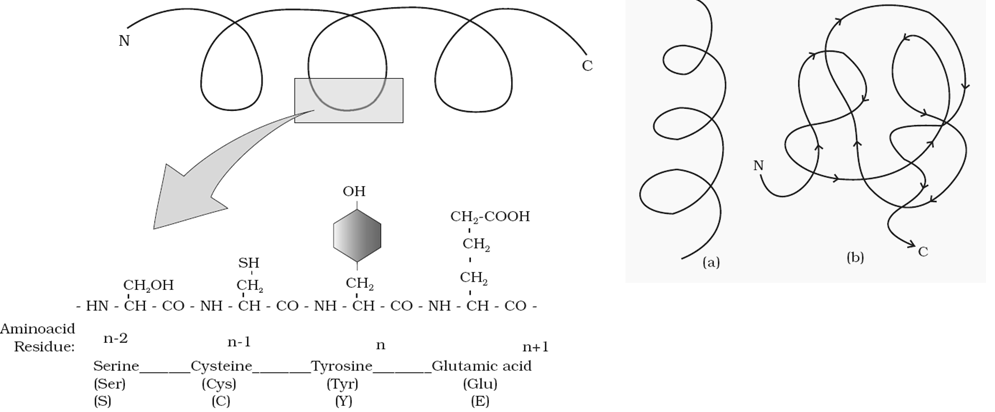
Structure of Proteins –
| Primary structure | Secondary structure | Tertiary structure | Quaternary structure | |
|
|
The Tertiary structure may result from further folding and coiling and may be stabilized by S-S (disulplhide) bond, Hydrophobic bonds and Ionic bonds.
This gives us a 3- dimensional view of a protein. Tertiary structure is necessary for many biological activities of proteins.
|
|
|
Functions of Proteins –
- Formation of cells and tissues for growth
- Repairing of tissues
- Formation of hormones
- For muscle contraction (eg. Actin, Myosin)
- Formation of enzymes
- Helps in blood clotting
- For transport (e.g. Haemoglobin, transferrin)
- For defense against infections (antibodies).
- Form hereditary material – Nucleoproteins
- For storage (e.g. Myoglobin and Ferritin)
- For support (e.g. Collagen and Elastin)
Tests for Proteins –
1) Biuret Test –
2) Xanthoproteic Test –
3) Millon’s Test
6. NUCLEIC ACIDS (DNA & RNA)
Chemical Structure of Nucleic Acid
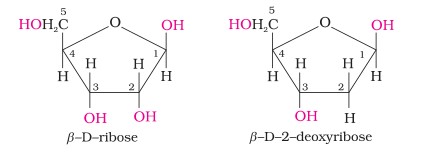
- Complete hydrolysis of DNA (or RNA) yields a pentose sugar, phosphoric acid and nitrogen containing heterocyclic compounds (called bases). In DNA molecules, the sugar moiety is β-D-2-deoxyribose whereas in RNA molecule, it is β-D-ribose.
- Sugar Molecule
DNA contains four bases viz., adenine (A), guanine (G), cytosine (C) and thymine (T). RNA also contains four bases, the first three bases are same as in DNA but the fourth one is uracil (U).
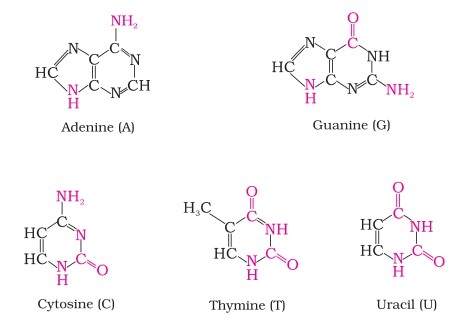
| Nucleic Acid | Purines | Pyrimidines |
| DNA | Adenine and Guanine | Cytosine and Thymine |
| RNA | Adenine and Guanine | Cytosine and Uracil |
NUCLEOSIDES
The combination of pentose sugar with nitrogenous bases (Purines or pyrimidines) is called nucleoside.
i. Nucleosides of purines
Adenine – Adenosine
Guanine – Guanosine
ii. Nucleosides of pyrimidines
Cytosine – Cytidine
Thymine (in DNA) or Uracil (in RNA) – Thymidine or Uridine
NUCLEOTIDES
Phosphate ester of a nucleoside is called nucleotide. Each nucleotide consists of a nitrogenous base, pentose sugar and one or more phosphate groups.
(i) Nucleotides of purines –
Adenosine + 1-phosphate group – Adenosine Monophosphate (AMP) or Adenylic acid
Guanosine + 1-phosphate group – Guanosine Monophosphate (GMP) or Guanylic acid
(ii) Nucleotides of Pyrimidines
Cytidine + 1-phosphate group – Cytidine Monophosphate (CMP)
Thymidine + 1-phosphate group – Thymidine Monophosphate (TMP) or Thymidylic acid
Uridine + 1-phosphate group – Uridine Monophosphate (UMP) or Uridylic acid
Formation of Dinucleotide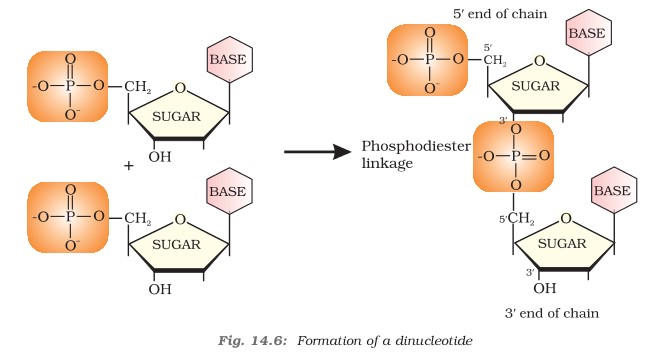
Nucleotides are joined together by phosphodiester linkage between 5′ and 3′ carbon atoms of pentose sugar. The chain of nucleic acid is abbreviated from 5′-3′.
Formation of Polynucleotide
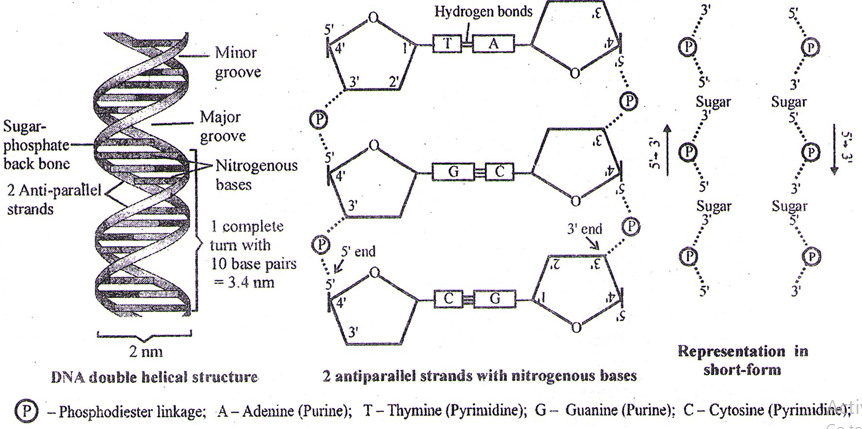
Double Helical Structure of DNA
To explain base equivalence (A/T, G/C) and other properties of DNA, Watson and Crick (1953), base on X-ray diffraction studies, proposed double helical structure of DNA.
- The diameter of double helical structure of DNA is 2 nm (20Å)
- The distance covered in 1 complete turn (pitch) = 3.4 nm (334 Å)
- Total base pairs in 1 complete turn = 10 (in B-type DNA)
- Total distance between 2 base – pairs = .34 nm (3.4 Å)
Based on number of base pairs and right or left handedness of helical, the DNA can be of the following main types
| Types of DNA | Type of Helices | No. of Base pairs per turn |
| A – Type | Rt. Handed | 11 |
| B – Type | Rt. Handed | 10 |
| C – Type | Rt. Handed | 9 1/3 |
| D – Type | Rt. Handed | 8 |
| Z – Type | Lt. Handed | 12 (6-nucleotides) Repeating unit dinucleotide |
Difference between DNA and RNA
| DNA | RNA |
| Double stranded | Generally single stranded |
| Sugar-Deoxyribose | Sugar-Ribose |
| Pyrimidines – Cytosine and Thymine | Pyrimidines – Cytosine and Uracil |
| Cytosines are equal to Guanines | Cytosines are not equal to Guanines (being single stranded) |
| Base pairs in millions | Base pairs usually 100 to 5000 |
Concept of Metabolism and Living state
- All the biomolecules have a turnover, which means, they are constantly being changed into some other biomolecules and also made from some other biomolecules, through chemical reactions.
- The living state is a non-equilibrium steady state to be able to perform work, and hence living process is a constant effort to prevent falling into equilibrium.
7. ENZYMES
- They act as catalysts in biological reactions and are, therefore, called Biocatalysts. Also known as metabolic regulators.
- Buchner, a German biochemist, discovered enzymes in yeast, and stated that living nature of the cells is not essential for the enzymatic activity.
- Kuhne coined the term ‘enzyme’ (means in yeast).
- J. Sumner isolated first enzyme (urease) in pure crystalline form from Jack beans.
Properties of enzymes
- Almost all enzymes are proteins. There are some nucleic acids that behave like enzymes. These are called ribozymes.
- Enzymes are oligodynamic, i.e. they are required in small amounts.
- Enzymes reduces the amount of activation energy required to start the chemical reaction.
- They can not change equilibrium point of a reversible reaction.
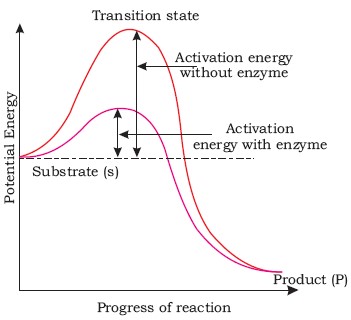
Active site
- It is a specific region in the structure of enzyme to which a substrate (reactant) binds. The active site has a specific type and number of amino acids. It is a small site and covers nearly 5% of the total area in most of the enzymes.
Turn over number
- The number of substrate molecules which can be catalyzed by a single molecule of an enzyme in a unit time. A maximum turn over number (36-milion mol./minute) is of Carbonic anhydrase. The turn over number depends up to the number of active sites.
Theories about enzymatic action
- Emil Fisher’s Lock and key hypothesis – According to this theory the active site of enzyme acts a as a lock while substrate acts as a key. Fisher believed that active site is rigid or non-flexible.
- Koshland Induced fit theory – According to this theory, the active site, is flexible, i.e., a change can be induced by the substrate, in the configuration of the active site.
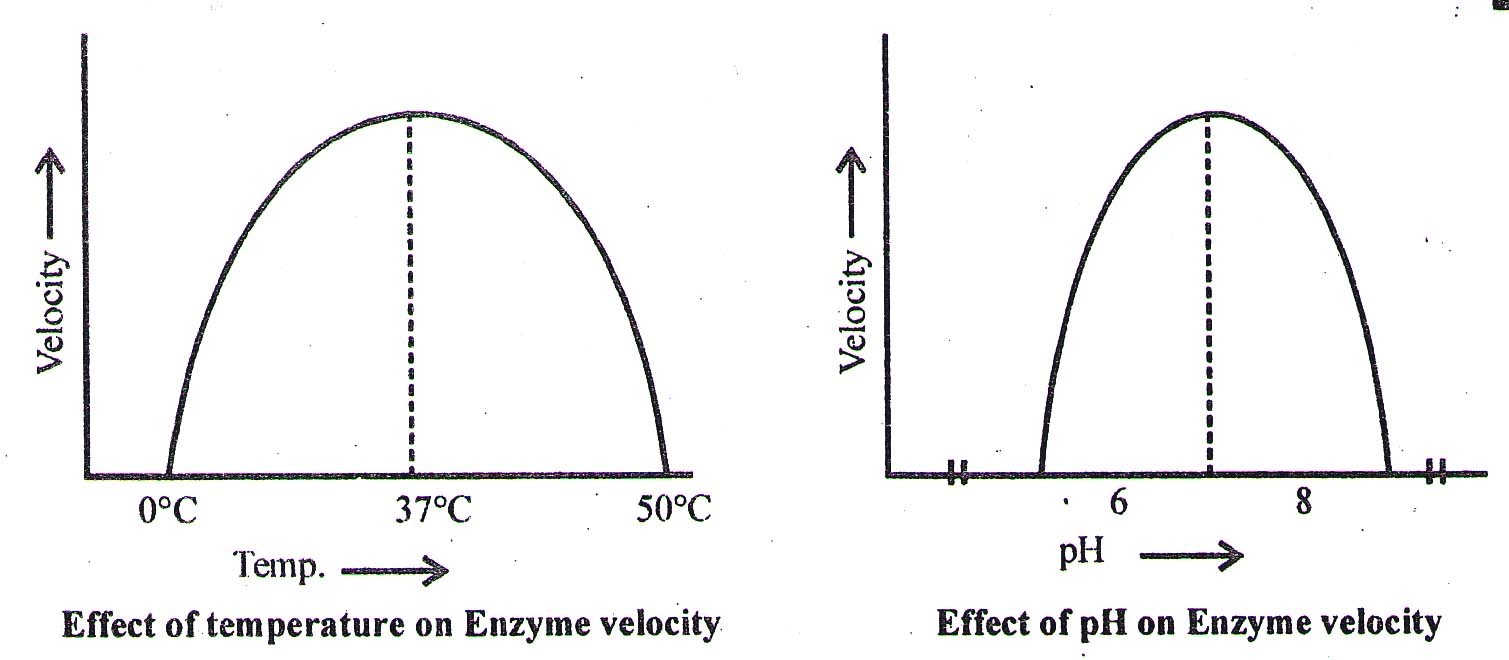
Effect of pH and Temperature on enzyme action
- Enzymes are pH specific, substrate specific and temperature specific. The optimum temperature for enzymatic action is ∼ 37°C. At 0°C the enzymes are inactivated while at 60°C or above most of the enzymes are denatured and their enzymatic activity is lost.
- The optimum pH for most of the enzymes is 7.
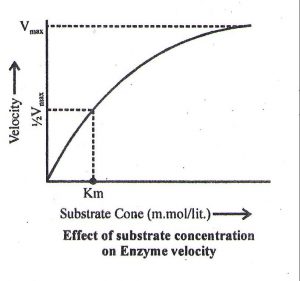
Effect of substrate concentration on the enzyme activity
- With the increase in concentration of substrate the enzymatic velocity also increases. At a certain value all the active site of the enzyme molecules are saturated. Increase in substrate concentration does not increase the velocity of the enzymatic reaction.
- Higher is the affinity of an enzyme for a substrate, lower is the Km value.
- Km value lies in between 10–1 to 10–6M
- Km value is independent of enzyme and substrate concentration
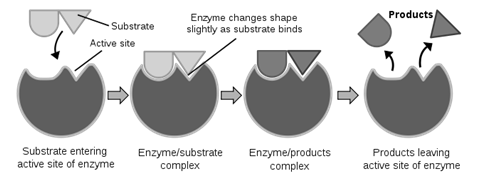
Nature of Enzyme Action
Each enzyme (E) has a substrate (S) binding site in its molecule so that a highly reactive enzyme-substrate complex (ES) is produced. This complex is short-lived and dissociates into its product(s) P and the unchanged enzyme with an intermediate formation of the enzyme-product complex (EP).
The formation of the ES complex is essential for catalysis.
The catalytic cycle of an enzyme action can be described in the following steps:
- First, the substrate binds to the active site of the enzyme, fitting into the active site.
- The binding of the substrate induces the enzyme to alter its shape, fitting more tightly around the substrate.
- The active site of the enzyme, now in close proximity of the substrate breaks the chemical bonds of the substrate and the new enzyme- product complex is formed.
- The enzyme releases the products of the reaction and the free enzyme is ready to bind to another molecule of the substrate and run through the catalytic cycle once again.
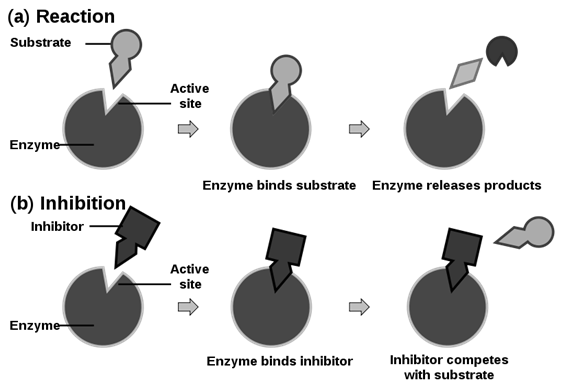
Inhibition of enzymatic action
1. By competitive inhibition
- A chemical, similar in configuration to the substrate, competes for the active site of the enzyme. E.g. Malonate (malonic acid) competes with Succinate (Succinic acid) for the active site of Succinate dehydrogenase. The malonate is the competitive inhibitor for the synthesis of Fumarate (Fumaric acid) from succinic acid, in this reaction.
The methanol toxicity is similarly averted by ethanol, a competitive inhibitor, for the enzyme alcohol dehydrogenase.
- Such competitive inhibitors are often used in the control of bacterial pathogens.
- Antibiotics containing sulpha drugs are similar in structure to PABA (para-aminobenzoate), a substance essential for the growth of may pathogenic (disease-causing) bacteria.
- No change in Vmax, but Km value increases.
2. By non-competitive inhibition
- Here the inhibitor does not compete with the substrate for active site. It binds with the enzyme at the site other than active site.
- The inhibitor changes the configuration of the active site or the 3-D shape of the enzyme. E.g., Cyanides stop the functioning of the respiratory enzymes by binding with the Iron fo the prosthetic group. Ions of Heavy metals (like Hg, Ag and Cu) combine with the disulphide/thiol group and break that to change the 3-D shape of the enzyme, or denature it.
- No change in Km value but Vmax decreases.
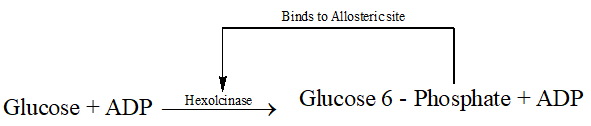
3. Allosteric inhibition (Feed back inhibition)
- Allosteric enzymes have Allosteric site with the active site. The modulator binds to this allosteric site and changes the rate of chemical reaction. The inhibitor in such reactions is one of the product of a long chain of enzymatic reaction and hence this inhibition is also known as feed back inhibition e.g. Hexokinase.
Classification of enzymes
- Enzymes are named according to the classification designed by Enzyme Commission (EC) of the IUPAC (International Union of pure and applied chemistry). This classification is based on the type of reaction which they catalyze.
- Enzymes are divided into 6 classes each with 4-13 subclasses and named accordingly by a four-digit number.
| Enzyme | Types of reaction | Example |
| Oxidoreductases/dehydrogenases | Enzymes which catalyse oxido-reduction between two substrates S and S’ S reduced + S’ oxidized → S oxidized + S’ reduced |
Oxidase (like Cytochrome oxidase), dehydrogenase (like Alcohol dehydrogenase)
|
| Transferases | Enzymes catalyzing a transfer of a group, G (other than hydrogen) between a pair of substrate S and S’ S – G + S’ → S + S’ – G |
Phosphorylase, transaminase and hexokinase
|
| Hydrolases | Enzymes catalyzing hydrolysis of ester, either, peptide, glycosidic, C-C, C-halide or P-N bonds. | Amylase, urease, lipase
|
| Lyases | Enzymes that catalyse removal of groups from substrates by mechanisms other than hydrolysis leaving double bonds.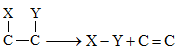 |
Decarboxylases, Deaminases |
| Isomerases | Includes all enzymes catalyzing inter-conversion of optical, geometric or positional isomers. | Aldolases |
| Ligases | Enzymes catalyzing the linking together of 2 compounds. It catalyse joining of C – O, C – S, C- N, P- O etc. bond | DNA and RNA ligases. |
Co-factors
- Enzymes composed of one or several polypeptide chains. However, there are a number of cases in which non-protein constituents are called cofactors
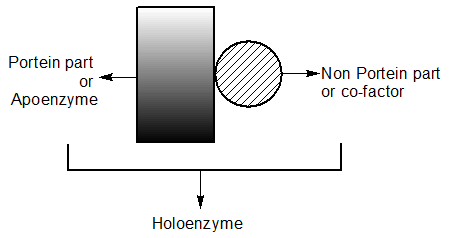
| Co-Factor | ||
| Prosthetic Group | Co-enzyme | Metal ions |
|
|
|








